REVIEW PAPER
Advanced methods of bacteriological identification in a clinical microbiology laboratory
1
Chair and Department of Medical Microbiology, Medical University, Lublin, Poland
Corresponding author
Magdalena Elwira Żukowska
Chair and Department of Medical Microbiology, Medical University of Lublin, Chodźki 1, 20-093, Lublin, Poland
Chair and Department of Medical Microbiology, Medical University of Lublin, Chodźki 1, 20-093, Lublin, Poland
J Pre Clin Clin Res. 2021;15(2):68-72
KEYWORDS
TOPICS
ABSTRACT
Introduction and objective:
Conventional, culture-based methods of bacterial identification and drug-susceptibility testing are considered the gold standard in medical microbiology. In recent years, classical microbiological methods have been supplemented with modern analytical and molecular methods. The aim of the review was to discusses the methods which have been permanently adapted to bacteriological microbiological diagnostics.
Abbreviated description of the state of knowledge:
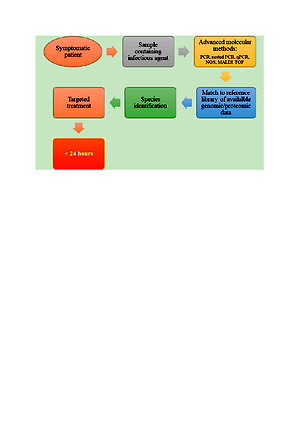
Currently, PCR, as well as other nucleic acid amplification tests and sequencing techniques, are part of the standard repertoire of microbiological diagnostics. With regard to the quality and speed of pathogen identification, the introduction of mass spectrometry techniques into routine microbiological diagnostics work-up has been revolutionary. Within a short time in many laboratories, Matrix-Assisted Laser Desorption/Ionisation – Time of Flight Mass Spectrometry (MALDI TOF MS) systems have almost completely replaced conventional biochemical pathogen identification.
Conclusions:
Microbiological diagnostics is an indispensable element of a targeted therapy. The techniques used in the laboratory depend primarily on the laboratory’s apparatus, the costs of the analysis, as well as the sensitivity and specificity of a method. However, regardless of the culture-based methods universality, advanced techniques have permanently established themselves in diagnostics. Confident information about the detected organism and treatment possibilities in a combination with the clinical context are conducive to successful therapy. Although modern methods still require validation and close collaboration between clinicians, microbiologists and bioinformaticians, these methods, once deemed to be the future, have already arrived.
Conventional, culture-based methods of bacterial identification and drug-susceptibility testing are considered the gold standard in medical microbiology. In recent years, classical microbiological methods have been supplemented with modern analytical and molecular methods. The aim of the review was to discusses the methods which have been permanently adapted to bacteriological microbiological diagnostics.
Abbreviated description of the state of knowledge:
Currently, PCR, as well as other nucleic acid amplification tests and sequencing techniques, are part of the standard repertoire of microbiological diagnostics. With regard to the quality and speed of pathogen identification, the introduction of mass spectrometry techniques into routine microbiological diagnostics work-up has been revolutionary. Within a short time in many laboratories, Matrix-Assisted Laser Desorption/Ionisation – Time of Flight Mass Spectrometry (MALDI TOF MS) systems have almost completely replaced conventional biochemical pathogen identification.
Conclusions:
Microbiological diagnostics is an indispensable element of a targeted therapy. The techniques used in the laboratory depend primarily on the laboratory’s apparatus, the costs of the analysis, as well as the sensitivity and specificity of a method. However, regardless of the culture-based methods universality, advanced techniques have permanently established themselves in diagnostics. Confident information about the detected organism and treatment possibilities in a combination with the clinical context are conducive to successful therapy. Although modern methods still require validation and close collaboration between clinicians, microbiologists and bioinformaticians, these methods, once deemed to be the future, have already arrived.
Żukowska ME. Advanced Methods of Bacteriological Identification in Clinical Microbiology Laboratory. J Pre-Clin Clin Res. 2021; 15(2): 68–72. doi: 10.26444/jpccr/134646
REFERENCES (27)
2.
Lau SKP, Teng JLL, Woo PCY. Bacterial Identification Based on Universal Gene Amplification and Sequencing. In: Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. 3rd ed. Springer Nature Switzerland; 2018. p. 1–2.
3.
Hou TY, Chiang-Ni C, Teng SH. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J Food Drug Anal. 2019; 27(2): 404–414. doi: 10.1016/j.jfda.2019.01.001.
4.
Boyles TH, Wasserman S. Diagnosis of bacterial infection. South African Medical Journal 2015; 105(5): 419. doi: 10.7196/SAMJ.9647.
5.
Bunn TW, Sikarwar AS. Diagnostics: Conventional Versus Modern Methods, Journal of Advances in Medical and Pharmaceutical Sciences 2016, 8(4): 1–7. https://doi.org/10.9734/JAMPS/....
6.
Szewczyk EM. Podłoża, próby i metody diagnostyczne. In: Szewczyk EM. Diagnostyka bakteriologiczna. 3rd edition. Warszawa: Wydawnictwo Naukowe PWN SA; 2019. p. 505–573.
7.
Żabicka D, Literacka E. Nowoczesne metody wykrywania i identyfikacji bakterii. Forum Zakażeń 2013; 4(1): 65–72. doi: dx.doi.org/10.15374/fz2013009.
8.
Kelley SO. New Technologies for Rapid Bacterial Identification and Antibiotic Resistance Profiling. SLAS Technol. 2017; 22(2): 113–121. doi: 10.1177/2211068216680207.
9.
Vila J, Gómez MD, Salavert M, et al. Métodos de diagnóstico rápido en microbiología clínica: necesidades clínicas. Enferm Infecc Microbiol Clin. 2017; 35(1): 41–46. doi: 10.1016/j.eimce.2017.01.014.
10.
Valones MA, Guimaraes RL, Brandao LA, et al. Principles and applications of polymerase chain reaction in medical diagnostic fields: a review. Braz J Microbiol. 2009; 40(1): 1–11. doi: 10.1590/S1517-83822009000100001.
11.
Yang S, Rothman R. PCR-based diagnostics for infectious disease uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004; 4(6): 337–348. doi: 10.1016/S1473-3099(04)01044-8.
12.
Idelevich E, Reischl U, Becker K. New microbiological techniques in diagnosis of bloodstream infections. Dtsch Arztebl Int. 2018; 115(49): 822–832. doi: 10.3238/arztebl.2018.0822.
13.
Persing DH, Tenover FC, Hayden RT, et al. Molecular Microbiology: Diagnostic Principles and Practice. 3rd edition. 2016.
14.
Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014; 27(4): 783–822. doi: 10.1128/CMR.00003-14.
15.
Szemraj M. Metody molekularne w diagnostyce mikrobiologicznej. In: Szewczyk EM. Diagnostyka bakteriologiczna. 3rd edition. Warszawa: Wydawnictwo Naukowe PWN SA; 2019. p. 475–490.
16.
Stratton CW. Tang YW. Interpretation and Relevance of Advanced Technique Results. In: Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. 3rd edition. Springer Nature Switzerland; 2018. p. 711–740.
17.
Massung RF, Slater K, Owens JH, et al. Nested PCR assay for detection of granulocytic ehrlichiae. J Clin Microbiol. 1998, 36(4): 1090–1095. doi: 10.1128/JCM.36.4.1090-1095.1998.
18.
Tondella ML, Talkington DF, Holloway BP, et al. Development and Evaluation of Real-Time PCR-Based Fluorescence Assays for Detection of Chlamydia pneumoniae. J Clin Microbiol. 2002; 40(2): 575–583. doi: 10.1128/JCM.40.2.575-583.2002.
19.
Cruz HL, et al. Evaluation of a Nested-Pcr for Mycobacterium Tuberculosis Detection in Blood and Urine Samples. Braz. J. Microbiol. 2011; 42(1): 321–329. doi: 10.1590/S1517-83822011000100041.
20.
Figueiredo da Costa Lima J, de Moraes Rego Guedes G, Falcao de Araújo Lima J, et al. Single-tube nested PCR assay with in-house DNA extraction for Mycobacterium tuberculosis detection in blood and urine. Revista da Sociedade Brasileira de Medicina Tropical. 2015; 48(6): 731–738. http://dx.doi.org/10.1590/0037....
21.
Alves da Silva D, de Pina LC, Rego AM, et al. Advances in the Diagnosis of Mycobacterium tuberculosis Infection. In: Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. 3rd edition. Springer Nature Switzerland; 2018. p. 101–136.
22.
Morganti S, Tarantino P, Ferraro E, et al. Next Generation Sequencing (NGS): A Revolutionary Technology in Pharmacogenomics and Personalized Medicine in Cancer. Adv Exp Med Biol. 2019; 1168: 9–30. doi: 10.1007/978-3-030-24100-1_2.
23.
Boers SA, Jansen R, Hays JP. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur J Clin Microbiol Infect Dis. 2019; 38(6): 1059–1070. doi: 10.1007/s10096-019-03520-3.
24.
Muldrew KL. Molecular Diagnostics of Sexually Transmitted Diseases: Bacterial, Trichomonas, and Herpes Simplex Virus Infections. In: Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. 3rd edition. Springer Nature Switzerland; 2018. p. 67–100.
25.
Hosseini S, Martinez-Chapa SO. Principles and mechanism od MALDI-ToF-MS analysis. In: Hosseini S, Martinez-Chapa SO. SpringerBriefs in Applied Sciences and Technology; 2017. p. 1–19. doi: 10.1007/978-981-10-2356-9_1.
26.
Singhal N, Kumar M, Kanaujia PK, et al. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015; 6: 791. https://doi.org/10.3389/fmicb.....
27.
Niu S, Chen L. Molecular Detection and Characterization of Carbapenem-Resistant Enterobacteriaceae. In: Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. 3rd edition. Springer Nature Switzerland; 2018. p. 165–186.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.